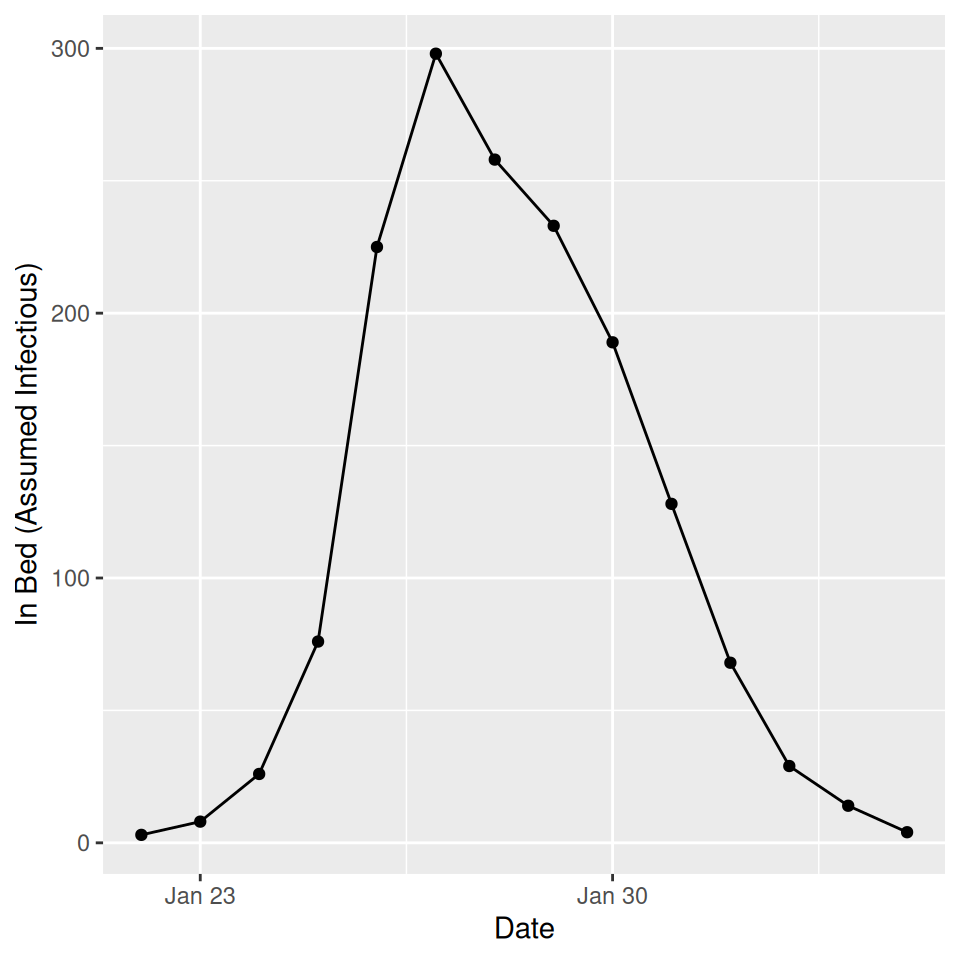
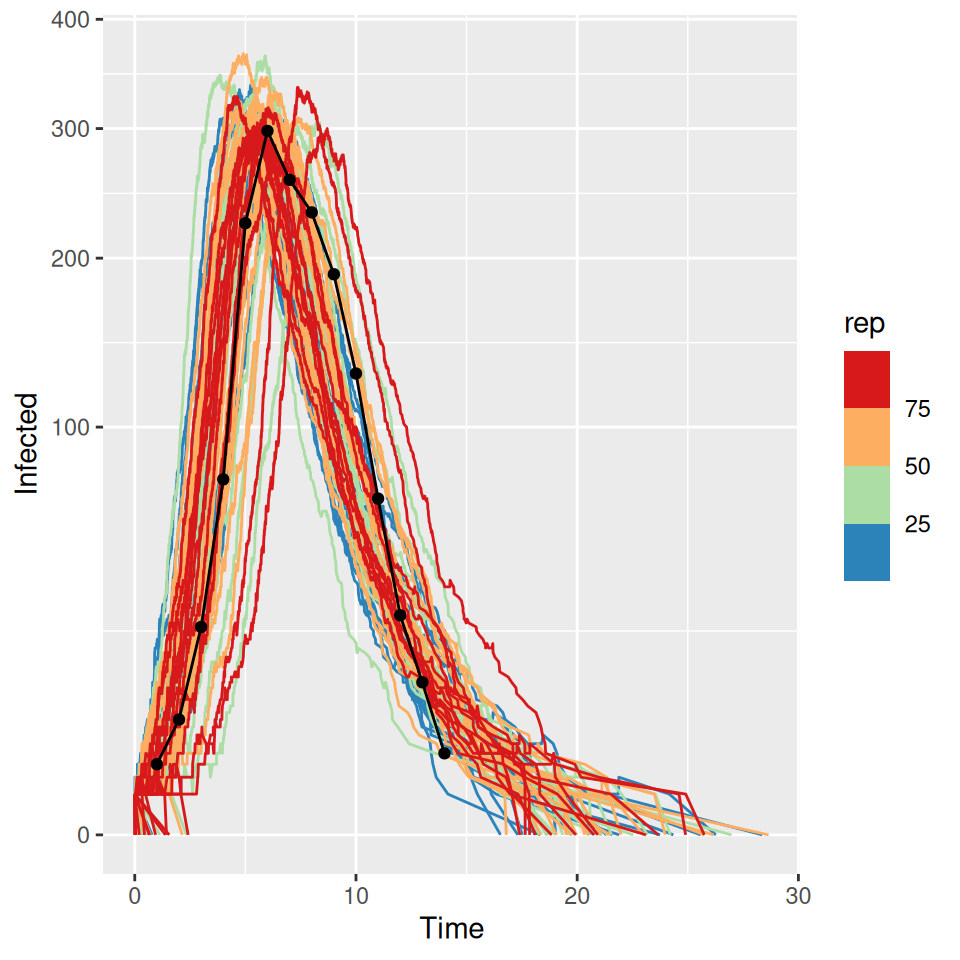
For the standard \(SEIR\) model with constant rates of progression through the exposed and infectious compartments an \(R_0\) of \(\sim 4.0\) gives a rough calibration to the observed data (black points and lines):
SEIR_gamma <- function(t,y,theta) {
x_i = integer(2);
beta = theta[1];
TE = theta[2];
TI = theta[3];
x_i[1] = theta[4];
x_i[2] = theta[5];
I_tot=0;
N_tot=0;
dy_dt = numeric(2+x_i[1]+x_i[2]);
for(k in (1+x_i[1]+1):(1+x_i[1]+x_i[2])){I_tot = I_tot + y[k];}
for(k in 1:(2+x_i[1]+x_i[2])){N_tot = N_tot + y[k];}
lambda = beta*I_tot/N_tot
#Susceptibles
dy_dt[1] = - (lambda) * y[1];
#First stage of Exposed (but not yet infectious)
dy_dt[2] = lambda * y[1] - (x_i[1]/TE) * y[2];
#If shape parameter > 1 update internal exposed stages
if(x_i[1]>1){
for(k in 3:(1+x_i[1]))
{dy_dt[k] = (x_i[1]/TE) * y[k-1] - (x_i[1]/TE) * y[k];}
}
#First infectious stage
dy_dt[(1+x_i[1]+1)] = (x_i[1]/TE)*y[(1+x_i[1])] - (x_i[2]/TI)*y[(1+x_i[1]+1)];
#If more than one infectious stage, run through internal stages
if(x_i[2]>1)
{
for(k in (3+x_i[1]):(1+x_i[1]+x_i[2]))
{dy_dt[k] = (x_i[2]/TI) * y[k-1] - (x_i[2]/TI) * y[k];}
}
# Final absorbing recovered stage
dy_dt[(1+x_i[1]+x_i[2]+1)] = (x_i[2]/TI)*y[(1+x_i[1]+x_i[2])];
return(list(dy_dt))
}
det_SEIR <-function(R0,TE,TI,shape_E,shape_I,vacc)
{
N0 = 763.0
theta = numeric(5)
beta = R0/TI
s0 = (1-vacc)*(N0-1)
e0 = 1
i0 = 0
r0 = vacc*(N0-1)
y0 = numeric(2+shape_E+shape_I);
y0[1] = s0;
y0[2] = 1
y0[2+shape_E+shape_I] = r0
theta[1] = beta;
theta[2] = TE;
theta[3] = TI;
theta[4] = shape_E
theta[5] = shape_I
y<-lsoda(y0,seq(0,30,1),SEIR_gamma,theta)
#y0 = y[dim(y)[1],2:(3+shape_E+shape_I)]
#y<-lsoda(y0,seq(0,prevacc*364,1),SEIR_gamma,theta)
# return total number infected
if(shape_E==1 & shape_I==1)
{return(y[,4])
}else if(shape_E==1 & shape_I > 1)
{
return(rowSums(y[,(2+1):(2+1+shape_I)]))
}else if(shape_E> 1 & shape_I == 1)
{
return(y[,(2+shape_E+1)])
}else
{
return(rowSums(y[,(2+shape_E+1):(2+shape_E+shape_I)]))
}
}
# Standard SEIR model (exponential latent, infectious periods)
expI <- det_SEIR(2.0,0.04,2.0,1,1,0.0)
det_calibrate <- tibble(time=1:14,var='R0 = 2',I=expI[1:14])
expI <- det_SEIR(4.0,0.04,2.0,1,1,0.0)
det_calibrate <- det_calibrate %>% bind_rows(tibble(time=1:14,var='R0 = 4',I=expI[1:14]))
expI <- det_SEIR(5.0,0.04,2.0,1,1,0.0)
det_calibrate <- det_calibrate %>% bind_rows(tibble(time=1:14,var='R0 = 5',I=expI[1:14]))
ggplot(det_calibrate,aes(x=time,y=I,col=var)) + geom_line() + geom_point() +
annotate('point',1:14,targetI[1:14]) + annotate('line',1:14,targetI[1:14])
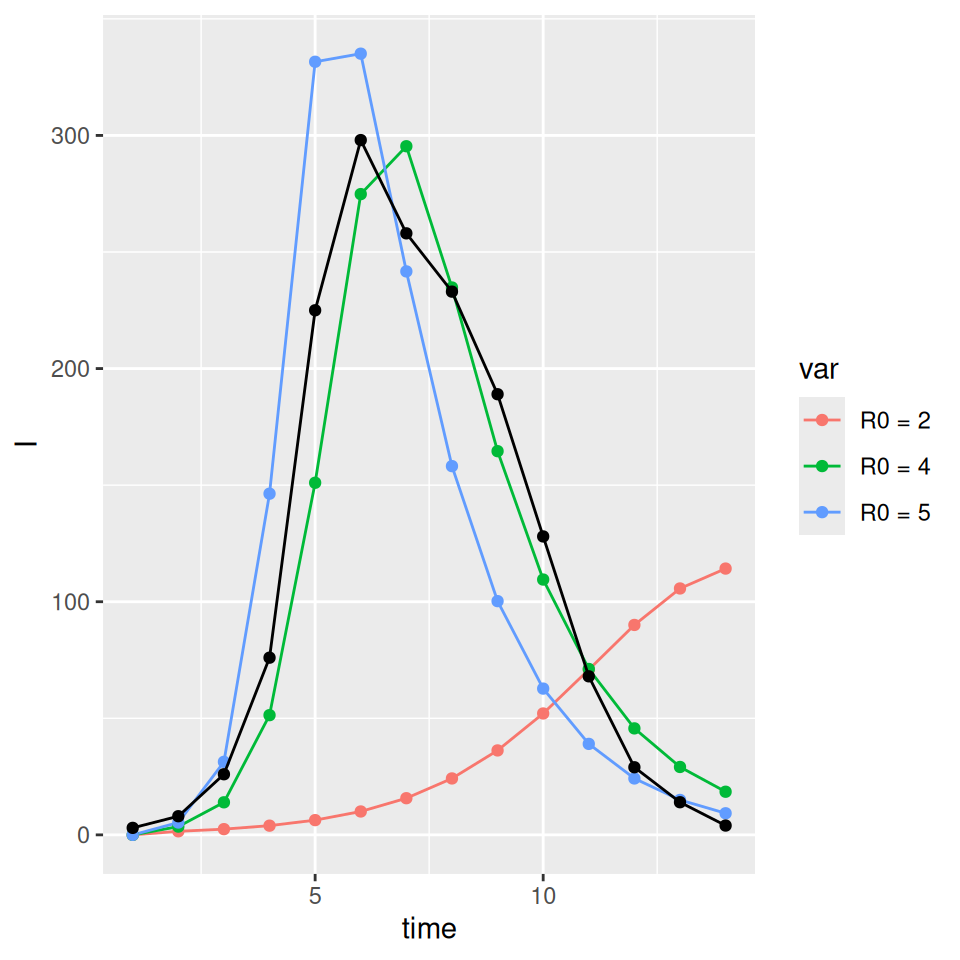
This rough calibration does not hold up if we now vary the distributional assumption for the latent and infectious periods. The supplied code has implemented the \(SEIR\) model with gamma distributed (strictly an Erlang distribution were the shape parameter is an integer) latent and infectious periods. If we vary the shape parameters for I and E we see that the peak prevalence and initial rate of exponential growth are very sensitive to changes in the distribution. Less dispersed distributions (i.e. higher shape parameters) have sharper epidemics with higher peak prevalence. The final size (and approximate value of \(R_0\)) on the other had are insensitive to these changes. Below we plot a set of three curves below for shape parameters = 10 illustrating that we now require a lower value of \(R_{0}\) to (roughly) match the data. The fit is not as good as I deliberately picked the average infectious and latent periods to give a reasonable fit with the exponential model…
# Standard SEIR model (gamma 10 latent, infectious periods)
expI <- det_SEIR(2.5,0.04,2.0,10,10,0.0)
det_calibrate <- tibble(time=1:14,var='R0 = 2.5',I=expI[1:14])
expI <- det_SEIR(2.75,0.04,2.0,10,10,0.0)
det_calibrate <- det_calibrate %>% bind_rows(tibble(time=1:14,var='R0 = 2.75',I=expI[1:14]))
expI <- det_SEIR(3.0,0.04,2.0,10,10,0.0)
det_calibrate <- det_calibrate %>% bind_rows(tibble(time=1:14,var='R0 = 3',I=expI[1:14]))
ggplot(det_calibrate,aes(x=time,y=I,col=var)) + geom_line() + geom_point() +
annotate('point',1:14,targetI[1:14]) + annotate('line',1:14,targetI[1:14])
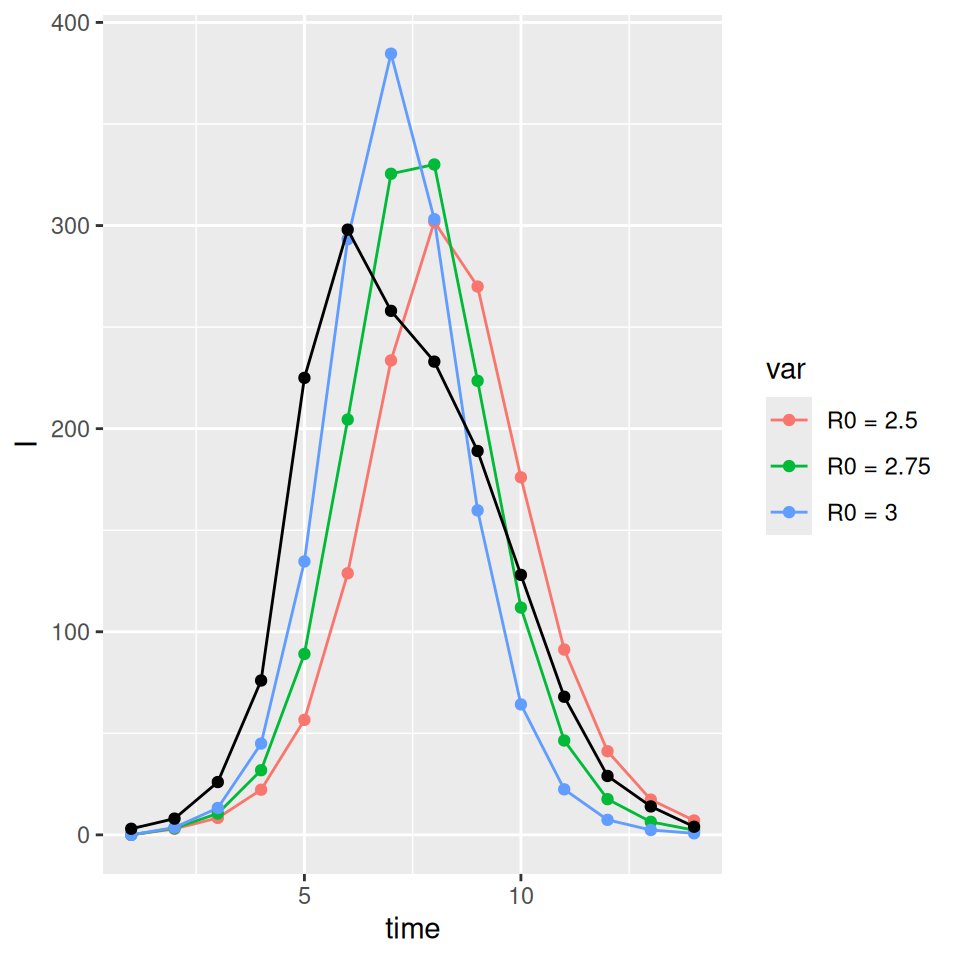
In practice the latent and infectious period distributions are not identifiable purely from a single epidemic curve. As this very simple comparison demonstrates calibrating our model based on the wrong assumption can lead to a significant under or over estimate of
\(R_{0}\).